Research
Biological macromolecular machines carry out a diversity of cellular processes. Their activities are often temporally and spatially coupled to one another, giving rise to new functions and forms of regulation. One preeminent example is the flow of genetic information from DNA sequence to protein product, which comprises many interacting players and interwoven reactions. Over the years a growing number of gene expression machines have been investigated in great detail. However, much less attention has been given to understanding the coordination and competition between these machines. We are building a multidisciplinary research program aiming to elucidate how cells execute the replication, transcription, translation, and degradation processes in a coordinated manner, and how these processes collectively respond to environmental changes. In other words, we are rethinking the central dogma of biology from a holistic point of view.
Projects
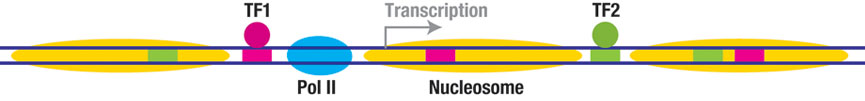 In eukaryotes, nucleosomes present significant barriers to Pol II transcription. In the meantime, numerous transcription factors (TFs) and DNA-binding proteins compete with the histones to bind to DNA, which profoundly affects nucleosome positioning and transcriptional output. We will build an experimental platform to directly visualize transcription through nucleosomes with different sets of TFs in real time. This platform will allow a global survey of transcriptional behavior across a large DNA sequence space. We will then use this platform to investigate the roles of chromatin remodelers, histone variants, and epigenetic modifications in transcriptional regulation.
In eukaryotes, nucleosomes present significant barriers to Pol II transcription. In the meantime, numerous transcription factors (TFs) and DNA-binding proteins compete with the histones to bind to DNA, which profoundly affects nucleosome positioning and transcriptional output. We will build an experimental platform to directly visualize transcription through nucleosomes with different sets of TFs in real time. This platform will allow a global survey of transcriptional behavior across a large DNA sequence space. We will then use this platform to investigate the roles of chromatin remodelers, histone variants, and epigenetic modifications in transcriptional regulation.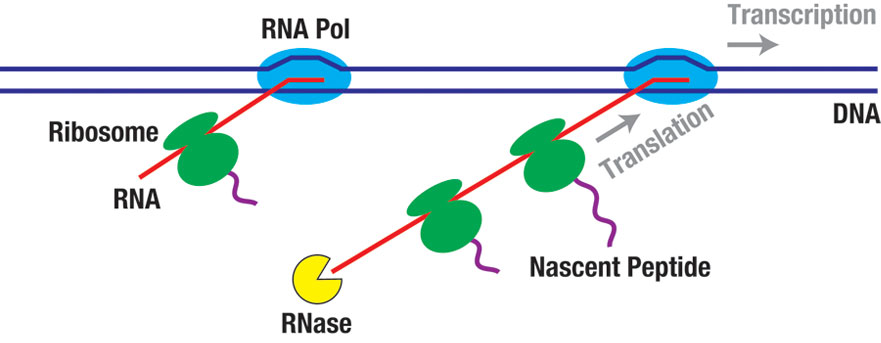 Describing the life cycle of RNAs in the cell is essential for understanding the dynamics of gene expression. At least a fraction of RNAs are co-transcriptionally degraded. In bacteria, ribosomes can bind to nascent transcripts and start translation before RNA synthesis is completed. The crosstalk between transcription, translation, and RNA degradation is a fascinating problem that is not well understood. We are developing single-molecule assays to study how RNA polymerase, the ribosome, and ribonucleases physically interact with each other. We will also leverage the power of high-throughput sequencing technology to characterize the interplay between these principal gene expression machines on a systems level.
Describing the life cycle of RNAs in the cell is essential for understanding the dynamics of gene expression. At least a fraction of RNAs are co-transcriptionally degraded. In bacteria, ribosomes can bind to nascent transcripts and start translation before RNA synthesis is completed. The crosstalk between transcription, translation, and RNA degradation is a fascinating problem that is not well understood. We are developing single-molecule assays to study how RNA polymerase, the ribosome, and ribonucleases physically interact with each other. We will also leverage the power of high-throughput sequencing technology to characterize the interplay between these principal gene expression machines on a systems level.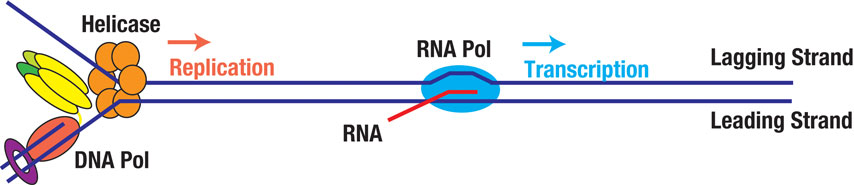 Replication and transcription machineries travel along the same DNA track and are doomed to collide. These collisions can cause DNA breakage and chromosomal recombination, and thus have been implicated in many types of cancers. We will use single-molecule and high-resolution cellular imaging approaches to monitor the sequence of molecular events before, during, and after replication-transcription collisions. We are especially interested in the fates of the replication fork and the transcription complex in this conflict. This study promises to shed new light on the mechanism of transcription-associated genome instability and inspire the development of novel strategies against cancer.
Replication and transcription machineries travel along the same DNA track and are doomed to collide. These collisions can cause DNA breakage and chromosomal recombination, and thus have been implicated in many types of cancers. We will use single-molecule and high-resolution cellular imaging approaches to monitor the sequence of molecular events before, during, and after replication-transcription collisions. We are especially interested in the fates of the replication fork and the transcription complex in this conflict. This study promises to shed new light on the mechanism of transcription-associated genome instability and inspire the development of novel strategies against cancer.